Top 10 Free Healthcare Datasets for Computer Vision
As anyone who works with computer vision models knows, the quality of a dataset directly impacts the performance and outcomes from training and production models. In this article, we’re sharing information and links to 10 of the best free, open-source datasets for healthcare computer vision models.
If you’re creating a medical imaging model, it’s crucial to get access to accurate and reliable medical imaging datasets.
This is especially important for medical imaging start-ups, who may not have the resources to build their own proprietary datasets. Open source medical imaging datasets can give artificial intelligence start-ups the data they need to get their first diagnostic model into production.
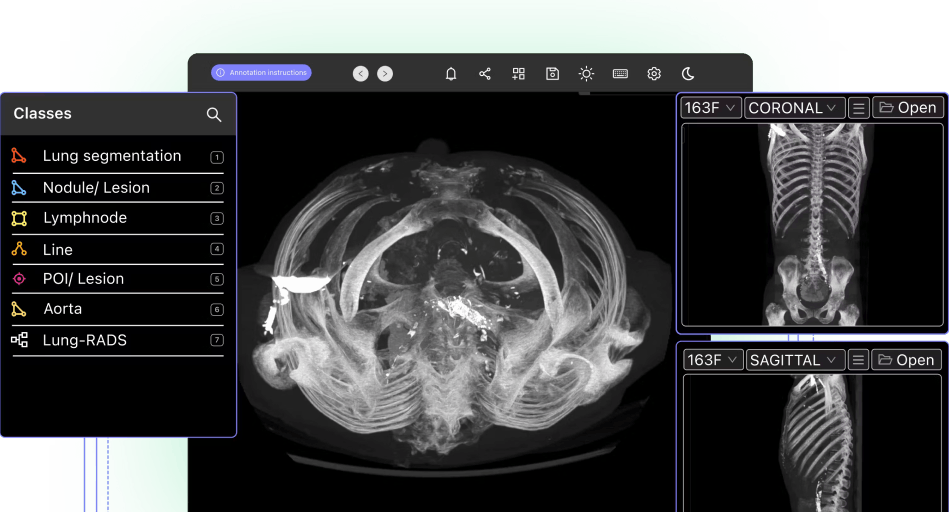
The Importance of Open-Source Medical Imaging Data
Open-source medical imaging datasets are useful because, in most cases, they’re ready to be labeled to create a training dataset. Images have been cleaned and cleansed, rarely contain identifiable patient data, and usually come with a wide range of metadata and other insights that are useful to medical researchers and healthcare providers.
Medical images and videos can come from numerous sources, such as microscopy, radiology, CT scans, MRI (magnetic resonance imaging), ultrasound images, X-rays (e.g. chest x-rays), and dozens of others. In most cases, these datasets are focused around a specific medical problem, such as cancer, COVID-19, scar tissue or other healthcare specialisms.
Depending on where the images have come from, they can come in a variety of formats. From standard image files (like PNG or JPG) to videos or medical specific file formats such as such as DICOM or NIfTI.
The value of these datasets can’t be understated, especially if you’re training a model for medical image analysis. Depending on the goals of your project, you might be able to use one of these public datasets to complete the project, or you might need access to proprietary medical imaging datasets once a model is trained to a sufficient level of accuracy.
For more information, check out our article on Best Practice for Annotating DICOM and NIfTI Files.
Top 10 Free Medical Datasets for Computer Vision
Now, let’s take a closer look at the top 10 free, open-source medical imaging datasets for computer vision models.
Here is an overview:
| Dataset | Type of Images | Number of Images | Key Features | Target Area |
| MedPix | Radiology images, with metadata | 59,000+ images | Metadata for each image; searchable by symptoms, diagnosis, organ, etc. | General medical imaging |
| Cancer Imaging Archive (TCIA) | CT, MRI, X-ray, PET, and histopathology images | Thousands of images | Includes metadata, treatment details, and research links | Cancer-related research |
| National COVID-19 Chest Imaging Database (NCCID) | Chest X-ray, CT, MRI | Varies by data release | Approved for research to support COVID-19 models | COVID-19 |
| COVID-19 Image Dataset | Chest X-ray images | 317 images (137 COVID-19) | Comparison of COVID-19, viral pneumonia, and healthy lungs images | COVID-19 detection |
| CT Medical Images | CT scan images | 475 images (69 patients) | Aimed at identifying textures and features for classification | Cancer research, CT analysis |
| OASIS Datasets | MRI brain scans | Thousands of images | Focus on Alzheimer's, mental illness, and neuroscience studies | Neuroimaging, brain health |
| Musculoskeletal Radiographs (MURA) | X-ray images of musculoskeletal scans | 40,561 images | Images labeled normal/abnormal by certified radiologists | Musculoskeletal imaging |
| re3data | Various medical imaging types | Millions of images | A library of datasets across medical domains | General medical research |
| NIH Deep Lesion Dataset | Radiology images (lesions) | Thousands of images | Deep lesions and accompanying metadata | Lesion detection, radiology |
| NIH Chest X-Ray Dataset | Chest X-ray images | 112,000+ images | Includes NLP labels for disease classification | Chest X-ray imaging |
MedPix
MedPix is a large-scale, open-source medical imaging dataset containing images from 12,000 patients, covering 9,000 topics and over 59,000 images. The dataset includes metadata from every image, and they’re organized according to where in the body the disease is (organ(s)), pathology, patient demographics, classification, and image captions.
Medical professionals can search the database according to symptoms, diagnosis, organs, image modality, description, keywords, and dozens of other choices.
MedPix is an open-source medical dataset hosted by The National Library of Medicine (NLM), at the Lister Hill National Centre for Biomedical Communications in Bethesda, MD.
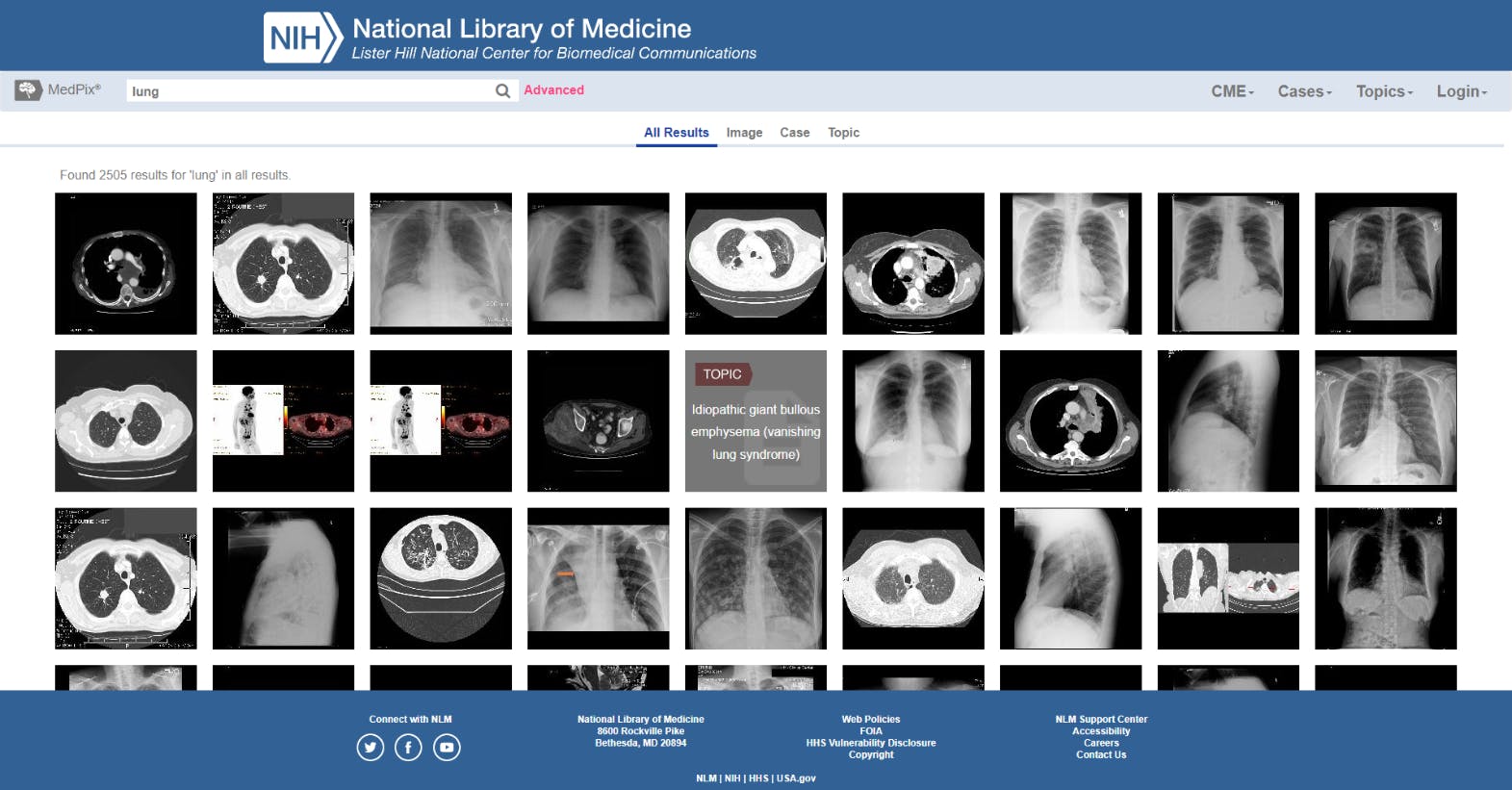
Examples of lung images in MedPix
The Cancer Imaging Archive (TCIA) Collections
Founded in 2011, The Cancer Imaging Archive (TCIA) is a National Institutes of Health (NIH) initiative created to support the cancer research community.
The high-quality TCIA open-source dataset includes thousands of “highly curated radiology and histopathology imaging, targeting prioritized research needs and supporting major NIH research programs.” Images include metadata, treatment details, pathologies, and even links to research and expert analysis, whenever available.
National COVID-19 Chest Imaging Database (NCCID)
Since the outbreak of the COVID-19 pandemic in March 2020, hospitals and health services worldwide have been collecting vast amounts of imaging data on this virus.
In the UK, the National Health Service (NHS) has collected a free, open-source database of COVID-19 patient chest images and X-rays. This database includes Chest X-Ray (CXR), Computed Tomography (CT), and Magnetic Resonance Images (MRI) from hospital patients across the UK.
Access to this database is approved through a medical board convened to ensure images will be used to improve patient outcomes and contribute to life-saving or life-enhancing treatments. One of the use cases the NHS anticipated is the dataset being used to develop, train, and support computer vision and AI models in the fight against Covid-19.
COVID-19 Image Dataset
On Kaggle, the open-source imaging dataset platform, you can also access a smaller dataset of Covid-19 patient Chest X-Rays.
This dataset includes 137 Covid-19 X-Ray images, plus others to compare against, including Viral Pneumonia and healthy chests/lungs. It contains 317 images, with 3 test directories and 3 training directories. All of the images come from researchers at The University of Montreal.
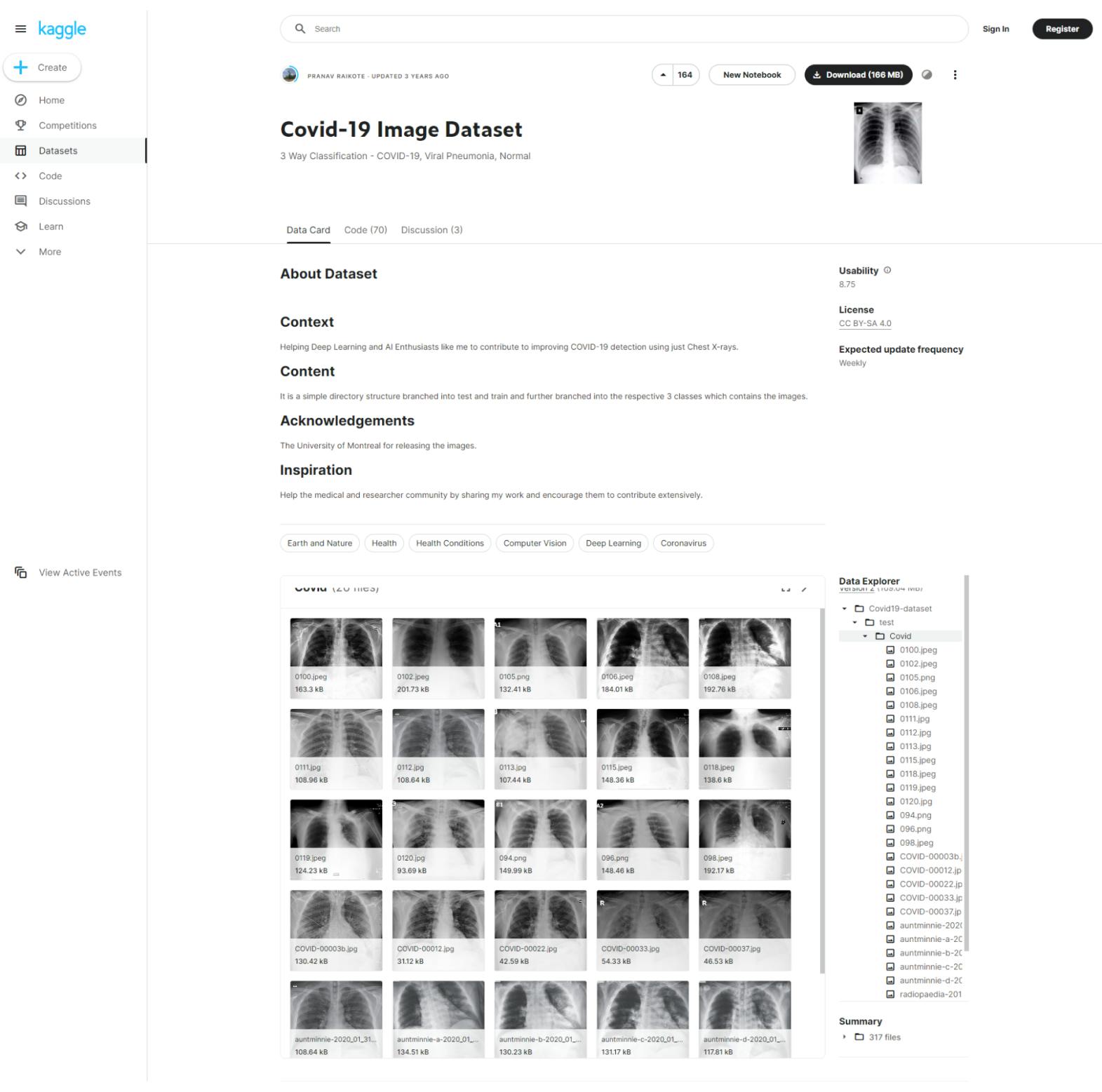
COVID-19 Dataset on Kaggle
CT Medical Images
Also on Kaggle is an open-source dataset that comes from CT images contained in The Cancer Imaging Archive (TCIA).
This dataset is very specific, containing images that come from the middle slice of CT images with the right age, modality, and contrast tags applied. It includes 475 images from 69 different patients.
The aim of this dataset “is to identify image textures, statistical patterns, and features correlating strongly with these traits and possibly build simple tools for automatically classifying these images when they have been misclassified.” It will prove useful to those building cancer-related computer vision models and researching this topic.
The OASIS Datasets
The OASIS (Open Access Series of Imaging Studies) Datasets contain four separate medical imaging datasets of brain scans. It includes thousands of images and patients.
These free open-source neuroimaging datasets are designed for medical professionals and medical providers studying a wide variety of brain-related healthcare issues. Making them ideal for training and testing computer vision algorithms that require neuroimaging data and metadata.
The OASIS Datasets are supported by National Institutes of Health (NIH) grants, and images come from a number of medical sources, including the Alzheimer’s Association, the James S. McDonnell Foundation, the Mental Illness and Neuroscience Discovery Institute, and the Howard Hughes Medical Institute (HHMI) at Harvard University.
Musculoskeletal Radiographs (MURA)
MURA (musculoskeletal radiographs) is an open-source musculoskeletal image database that started out as a Stanford University School of Medicine machine learning (ML) competition. Even though the competition is now closed, anyone can request access and download the dataset for the purposes of medical research and training machine learning models.
MURA contains 40,561 multi-view radiographic X-Ray images from 14,863 studies and 12,173 patients. Every image was collected from studies and patient scans where the images were labeled either normal or abnormal by board-certified radiologists from Stanford Hospital between 2001 and 2012.
re3data
re3data (Registry of Research Data Repositories) is a vast open-source library of medical imaging datasets. It’s like a Google or JStor of medical imaging datasets. re3data gives medical researchers and anyone training and testing medical imaging computer vision models access to millions of images and datasets, across dozens of medical specialisms.
Funding comes from the German Research Foundation (DFG), with support from other German institutions and libraries, such as the Karlsruhe Institute of Technology (KIT).
NIH Deep Lesion Dataset
The National Institutes of Health (NIH) Deep Lesion Dataset is an open-source dataset containing thousands of deep lesion images. It was created in 2018, and supported by NIH funding, and anyone can download the images through a simple Box folder.
NIH Chest X-Ray Dataset
This is a large-scale dataset from NIH, available through Kaggle, and offers a series of over 112,000 chest X-Ray images from more than 30,000 unique patients. It’s the largest known open-source chest X-Ray imaging dataset in the world, with images scoring a 90%+ accuracy, with the majority suitable for weakly-supervised computer vision learning.
Every image is supported by Natural Language Processing (NLP) so that labels align with disease classifications supplied by radiological reports. It’s an incredibly valuable resource for anyone conducting machine learning or computer vision modeling that requires chest X-ray images.
Medical Image Annotation with Encord
For those using a proprietary medical imaging dataset, these aren’t usually labeled images for the purposes of training a model. Equally, for most computer vision models, data scientists will want to label these images according to the goals of their model.
In this scenario, which is more common across the medical sector, clinical ops teams will need to get the dataset annotated by trained medical professionals before the dataset is ready to train a computer vision model.
Of course, annotators can more effectively deliver their work when they’ve got access to the right tools. Such as an annotation platform like Encord Annotate.
Encord streamlines collaboration between medical professionals, machine learning teams, and annotators. With Encord, you can accelerate the process of labeling medical imaging data, produce more accurate datasets and ultimately get your models into production more quickly.
Encord Annotate gives annotators access to a range of annotation types (including bounding boxes, human pose estimation, pixel-perfect auto-segmentation and object detection).
Encord has developed our medical imaging dataset annotation software in close collaboration with medical professionals and healthcare data science teams, giving you a powerful automated image annotation suite, fully auditable data, and powerful labeling protocols.

Frequently asked questions
Open-source medical imaging datasets provide high-quality, pre-processed data that can be used to train and test computer vision models. These datasets are often cleaned, anonymized, and come with detailed metadata, saving time and resources for medical imaging start-ups and researchers.
These datasets include a variety of imaging modalities such as X-rays, MRIs, CT scans, ultrasounds, and microscopy images. Many datasets focus on specific medical conditions, such as cancer, COVID-19, or neurological disorders.
Common file formats include standard image formats like PNG and JPG, as well as medical-specific formats such as DICOM (Digital Imaging and Communications in Medicine) and NIfTI (Neuroimaging Informatics Technology Initiative).
Cancer research: The Cancer Imaging Archive (TCIA)
COVID-19 studies: National COVID-19 Chest Imaging Database (NCCID)
Brain imaging: OASIS Datasets
Musculoskeletal analysis: MURA Dataset
Each dataset’s description in the article can guide you to the right choice.Annotation tools like Encord Annotate offer features such as pixel-perfect auto-segmentation, bounding boxes, and human pose estimation. These tools are designed for efficient collaboration between medical professionals and machine learning teams.
Encord Annotate simplifies and accelerates the annotation process, supports various annotation types, and provides powerful audit and labeling protocols. It is specifically tailored for medical imaging datasets, ensuring high-quality annotations for AI model training.
Yes, Encord can assist in automating annotation processes for clinical trial data through its innovative platform. By leveraging our tools and access to specialized annotators, you can streamline the annotation workflow, reduce manual efforts, and improve the overall efficiency of your data management tasks. This is particularly beneficial for clients looking to enhance their data processing capabilities.
Encord provides robust features for data annotation specifically designed for healthcare applications, including support for various data formats such as DICOM and NDPI. The platform allows users to annotate data independently or collaborate with third-party service providers, ensuring a streamlined workflow for medical data handling.
Encord provides a comprehensive annotation platform that is designed to assist healthcare customers with data labeling and enrichment. This includes curated samples of healthcare datasets, leveraging expert knowledge, and offering a catalog that categorizes datasets based on specific use cases and modalities.
Encord is designed to integrate various types of patient monitoring data, including telemetry feeds and video segments. The platform supports time series analysis, enabling clinicians to identify and annotate specific events, such as tachycardia or arrhythmias, within the monitoring data.
Encord provides robust support for integrating and managing both electronic health records and DICOM data. This allows users to leverage existing healthcare data efficiently while ensuring that all annotation and review processes are consolidated within a single platform.
Encord provides a range of features tailored for healthcare organizations, including compliance with regulatory pathways and tools to manage sensitive data effectively. The platform's capabilities are designed to support the unique demands of the healthcare industry.
Yes, Encord offers access to sample datasets, including a substantial open-source dataset in the medical space containing around 20,000 labeled frames. This allows users to test the platform's capabilities without needing to provide extensive custom data initially.
Yes, Encord is designed to integrate seamlessly with existing healthcare systems. This allows for enhanced data management and analysis workflows, ensuring that users can leverage their current tools while benefiting from Encord's advanced annotation capabilities.
Yes, Encord has capabilities that allow users to filter and access historical data efficiently. It enables radiologists to annotate data based on specific criteria, ensuring that the right data is routed to the appropriate projects for analysis.
Encord prioritizes patient privacy and security by not sharing identifiable patient data. The platform is designed to handle sensitive health information (PHI) responsibly, ensuring that all data is anonymized and secure during analysis.

