Contents
Step 1: Select and Divide the Datasets
Step 2: Prepare to Annotate with Multiple Blinds
Step 3: Establish the Image Annotation Protocol
Step 4: Practice Medical Image Annotation on a Handful of Samples
Step 5: Release the First Batch of Images for Labeling In Your Annotation Tool
Step 6: Release the Rest of the Data for Annotation and Implement Continuous Labeling Monitoring
Building a Tool for Medical Annotations Means Understanding Medical Professionals
Encord Blog
How to Structure QA Workflows for Medical Images
When building any computer vision model, machine learning teams need high-quality datasets with high-quality annotations to ensure that the model performs well across a variety of metrics.
However, when it comes to building artificial intelligence models for healthcare use cases, the stakes are even higher. These models will directly affect individual lives. They need to train on data annotated by highly skilled medical professionals who don’t have much time to spare. They’ll also be held to a high scientific and regulatory standard, so to get a model out of development and into production, ML teams need to train it on the best possible data with the best possible annotations.
That’s why every computer vision company–but especially those building medical diagnostic models– should have a quality assurance workflow for medical data annotation.
Structuring a quality assurance workflow for image annotation requires putting processes in place to ensure that your labeled images are of highest possible quality. When it comes to medical image annotation (whether for radiology modalities or any other use case) , there are a few additional factors to consider when structuring a QA workflow. If you take these considerations into account when building your workflow and have the framework for your workflow in place before you begin the annotation process, then you’ll save time at the later stages of model development.
Because medical image annotation requires medical professionals, annotation can be a costly part of building medical AI models. Having a QA workflow for image annotation before beginning model development can help a company to budget accordingly and reduce the risk of wasting an annotator’s time at the company’s expense.
Step 1: Select and Divide the Datasets
Medical models need to train on vast amounts of data. Your company will need to source high-quality training data while carefully considering the amount and types of data required for the model to perform well on its designated task. For instance, some tumors are rarer than others. However, the model needs to be able to classify rare tumors should it come across them “in the wild,” so the sourced data must contain enough examples of these tumors to learn to classify them accurately.
Before you begin building your QA workflow, a portion of the data needs to be separated from the total data collected. This fraction becomes the test data– the never-before-seen data that you’ll use after the training and validation stages to determine if your model meets the performance threshold required to be released into clinical settings. This data should not be physically accessible to anyone on the machine learning or data engineering teams because when the time comes to obtain regulatory approval, the company will have to run a clinical study, and doing so will require using untouched data that has not been seen by the model or anyone working on it. Ideally, this test data will be copied onto a separate hard drive and kept in a separate physical location so as to ease the burden of showing compliance during the regulatory approval process.
When building medical imaging models, you’ll also need to think carefully about the amount and types of annotations required to train the model. For instance, for those rare tumors, you need to decide how many examples need to be labeled, how often annotators will label them, and how annotators will categorize them.
Your company might source millions of mammograms or CT scans, but, in reality, medical professionals won’t have enough time to label all those pieces of data, so you’ll have to make a decision about how to schedule the annotation process..
To do so, you’ll have to decide on an amount of representative data and split that data into a training set and a validation set. However, before you split the data, you’ll also need to decide how many times each piece of data will be labeled. By computing a consensus, you ensure that you are not modeling a single annotator.
Step 2: Prepare to Annotate with Multiple Blinds
In medical imaging, single labeling is not sufficient. The images need multiple labels by different labellers just like scans need to be read by multiple doctors during clinical practice. In most European and North American countries, double reading is standard practice: each medical image is read by at least two radiologists.
At a minimum, your validation set will need to be double labeled. That means that different annotators will need to label the same piece of data. Furthermore, you may want to let annotators label the same data multiple times. By doing so, you can compute the inter and intra reader agreement. Of course, having multiple annotators is costly because these annotators are medical professionals– usually radiologists– with a significant amount of experience.
The majority of the data, say 80 percent, will fall into the training data set. The cost and time constraints associated with medical image annotation generally mean that the training data is often only single labeled, which enables the model to begin training more quickly at less cost.
However, the remaining data, which makes up the validation set, will be used to evaluate the model's performance after it completes its training. Most companies should aim to secure additional labels for the validation data. Having five or so annotators labeling each image will provide enough opinions to ensure that the predictions of the model are correct. The more opinions you have, the less the model will be biased towards a specific radiologist’s opinion and generalize better on unseen data.
This division of labor should be determined when setting up the annotation pipeline. The annotators don’t know how many times a piece of data is being labeled, and ideally the labeling is always completed blind. They shouldn’t talk to one another or discuss it. When working in a hospital setting, this confidentiality isn’t always guaranteed, but when working with a distributed group of radiologists, the double blind remains intact and uncompromised.
Step 3: Establish the Image Annotation Protocol
Now that you’ve collected and divided the data, you’ll need to establish the labeling protocol for the radiologists.
The labeling protocol provides guidelines for annotating “structures of interest”– tumors, calcifications, lymph nodes, etc.– in the images. The correct method for labeling these structures isn’t always straightforward, and there is often a trade-off between what is done in clinical routine and what is needed for training a machine learning.
For instance, let’s say you have a mass– a dense region is the breast for example. This mass can be round but it can also be star shaped. The annotator needs to know whether they should circle the mass or closely follow the outline. That decision depends on what the AI system will need to do in a clinical setting. If you’re training a system that only has to detect whether a mass is present, then a loose annotation might be sufficient. However, if the system is trying to discriminate between masses of different shapes, then you’ll likely need to segment it very carefully by following its exact outline. Often, irregular shaped masses tend to be signs of more aggressive cancers, so the machine will definitely need to be able to identify them.
Another example is calcification in medical imaging, which looks like salt and pepper noise on a medical image. How should that be annotated? A box around all the grains? A circle around each grain? A large bounding box means a compromise for the machine’s learning because it contains both calcifications and normal tissue, but it’s also unreasonable to ask doctors to annotate hundreds of small dots. You’ll need to detail what annotators should do in this situation in the labeling protocol. The same goes for encountering other objects– such as pacemakers and breast implants– in a scan. If annotators need to label these objects, then you must instruct them to do so.
A member of the machine learning team and someone with a clinical background should produce the labeling protocol together because different subject-matter experts think about these things differently. Remember that doctors don’t think about distinguishing a pacemaker from a tumor. They have years of experience and the ability to think critically, so to them it seems ridiculous that someone might mistake a pacemaker for a cancerous tumor. However, models can’t reason: they’ll only learn what the labels specifically point out to them in the medical images. Often, machine learning teams need to explain that to the radiologists. Otherwise, doctors might not understand why it matters if they leave a pacemaker unlabelled or circle an irregular shaped mass in one image and outline it in the next.
Be as explicit and exhaustive as possible. Annotation is a tedious and time consuming task, so labelers will understandably look for ways to cut corners unless you instruct them not to. Provide them with a labeling protocol manual that is precise but not overly lengthy. Include some pictures of good annotations and poor annotations as examples of what to do and what not to do. Then onboard them with a webinar session in which you share examples and demo the annotation platform so the labellers know what to expect and how to annotate within the platform.
Without a detailed labeling protocol, labellers may produce inconsistent labels. A common mistake is mixing up left and right when asked to annotate a specific structure, e.g. “label the left lung.” And loose annotations– circling rather than following the outline– often occur simply out of habit.
Step 4: Practice Medical Image Annotation on a Handful of Samples
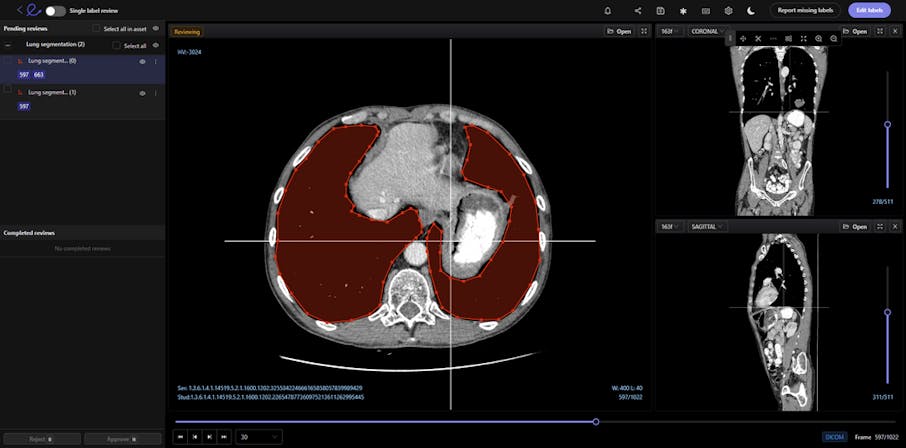
DICOM images contain a wealth of information that enables the best possible diagnosis of a patient. However, labeling volumetric images, such as CT or MRI scans, is challenging.
Encord’s DICOM Annotation Tool was designed in close collaboration with medical professionals, so unlike other existing DICOM viewers, it allows for seamless annotations and the building of a data pipeline to ensure data quality. Most existing data pipeline tools can’t represent the pixel intensities required for CT and MRI scans whereas our platform provides accurate and truthful displays of DICOMs. While some of our competitors convert DICOMs to other formats (e.g. PNGs, videos), we allow users to work directly on DICOMs, so that nothing is lost in conversion. By providing annotators with functions like custom windowing and maximum intensity projection, we’re enabling them to work similar to what they are used to in clinical practice, so that they can accurately assess images without the interference of shifting data quality.
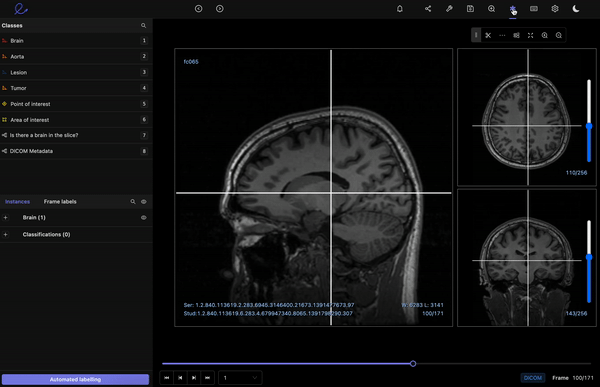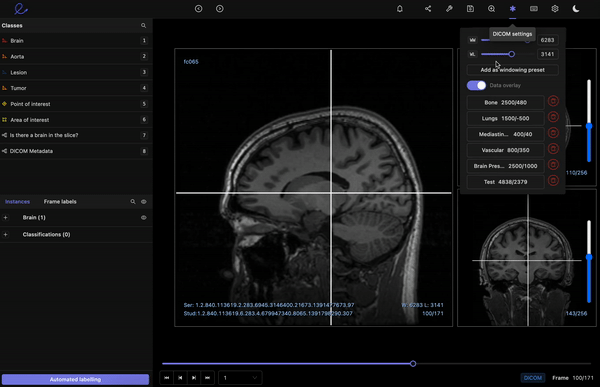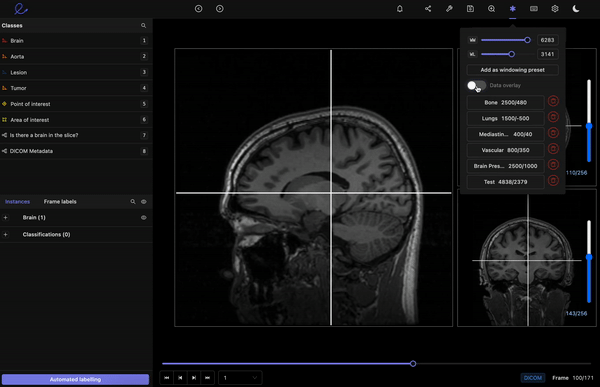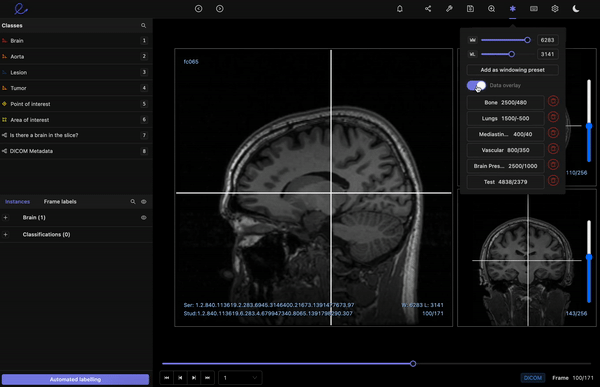
DICOM metadata in Encord
Volumetric images contains many slices and take a lot of time to investigate. Encord’s tool also supports maximum intensity projection in which users can collapse multiple slices into one plane layer, providing them with the opportunity to review the image from a different perspective– a perspective that might reveal findings otherwise missed. All of these features and more should help annotators better master the labeling protocol and produce high-quality medical image annotations more efficiently.
Providing the right tools will help your annotators do the best possible job in the least amount of time. However, regardless of the tools used, before deploying the training data for annotation, you should provide each radiologist a handful of samples to annotate and then have a meeting with them, either as a group or individually, to discuss how they think it went.
Work with a clinical expert to review that handful of samples to determine whether the labels achieve the high quality needed for training the machine learning models and algorithms. Compare the data samples to one another to determine whether one annotator performs significantly better or worse than the others. Consider questions like: Does one annotator label fewer structures than the others? Does another draw loose bounding boxes? Some variation is expected, but if one annotator differs significantly from the others, you may have to meet with them privately to align expectations.
Even with a labeling protocol, closely reviewing these handful of samples is essential for catching misalignments in thinking before releasing too much of the data. Remember, doctors think like doctors. If a patient's scan reveals 13 cancerous tumors in the liver, a doctor might only circle seven, because, in a clinical setting, that would be enough to know the patient has cancer and needs treatment. However, machine learning teams need to make sure that the doctor labels all 13 because the model will encounter those additional six and become penalized by the missing label. Missed annotations will make estimating the true model performance impossible, so machine learning teams need to help doctors understand why they need to perform exhaustive labeling, which is more time-consuming and differs from their routine clinical work.
Different annotators will have different thresholds for what they think should be annotated, so you’ll need the input of a clinical partner to determine what should have been annotated. Uncertainty always exists in medical image evaluation, so you’ll need to calibrate the annotators, telling them to be more or less sensitive in their thresholds.
Step 5: Release the First Batch of Images for Labeling In Your Annotation Tool
Once all the radiologists understand the labelling expectations, it’s time to release a first batch of images for annotation. For the first batch, release about a third of the data that needs to be annotated.
Set a timeline for completing the annotations. The timeline will depend on your company’s time constraints. For instance, if you are working towards attending a conference, you’ll need to train the model sooner rather than later, and you’ll want to shorten the timeline for annotations.
Someone from the machine learning team and the clinical partner should oversee and review the annotation process. That means you need to build time in for quality control. Reviewing annotations takes time, and, ideally, you’ll keep a record of each annotator’s labelling quality, so that you have monthly or weekly statistics that show how well each radiologist labelled an image compared to the ground truth or to a consensus.
When it comes to medical image annotation, establishing a ground truth requires finding information about the patients’ outcomes, which can be tricky. For instance, if three doctors read an image of a mass as non-cancerous, but a biopsy later revealed that the mass was cancerous, then the ground truth for that image is actually “cancerous.” Ideally, when you collect your data, you’ll receive clinical data along with the DICOM image that provides information about the patient’s treatment and outcomes post-scan, allowing for the establishment of a ground truth based on real-world outcomes.
Because Encord platform supports DICOM metadata, if the clinician and radiographer have collected this metadata, using Encord will enable you to seamlessly access important information about the patient’s medical condition, history, and outcomes.
In the case that no such clinical information is available, a consensus derived from annotations will have to serve as a proxy for the ground truth. A consensus occurs when a group of radiologists read the same scan, and arrive at an agreement, usually via a majority voting, about the finding. That finding then serves as the ground truth for the data.
However, in a clinical setting, doctors take different approaches to determining consensus. That’s why Encord’s platform provides a variety of features to help compute a consensus. It includes templates for maturity voting for any amount of annotators. It also features weighting so that a more experienced medical professional’s annotations would be given greater consideration than a more junior-level one. When disagreements about an image arise, Encord’s platform enables an arbitration panel in which the image is sent to an additional, more experienced professional to decide the consensus. Having a variety of approaches built into the platform is especially useful for regulatory approval because different localities will want companies to use different methods to determine consensus.
Within this part of the QA workflow, you should also build in a test of intra-rater reliability, in which each reviewer receives a set of data which contains the same image multiple times. The goal is to ensure that the rater performs consistently across time. This continuous monitoring of the raters answers important questions such as: Does the reviewer perform as well in the morning as in the evening? Does the reviewer perform poorly on the weekend compared to the weekdays?
Regulatory processes for releasing a model into a clinical setting expect data about intra-rater reliability as well as inter-rater reliability, so it’s important to have this test built into the process and budget from the start.
Step 6: Release the Rest of the Data for Annotation and Implement Continuous Labeling Monitoring
If the review of the first batch of annotations went well, then it’s time to release the rest of the data to the annotators. In general, if there’s a strict timeline or a concrete amount of data, a company will release the rest of the data in distinct batches and provide deadlines for labelling each batch. Otherwise, most companies will implement a continuous labeling stream. When a company has access to an ongoing flow of data from different manufacturers, a continuous labeling stream is the best strategy.
Continuous labeling streams require continuous labeling monitoring, and continuous labeling monitoring is great because it provides interesting and important insights about the labels and the data itself.
Encord’s DICOM annotation tool provides machine learning teams with access to important metadata that may have an impact on annotations and machine performance. DICOM data contains information about the state of the machine– its electrical current, X-ray exposure, angle relevant to the patient, surrounding temperature, and more.The team can also break the data broken down by country, costs, and manufacturer.
All of this information is important because it contributes to the appearance of the image, which means that the metadata has an impact on model performance. For instance, if labellers consistently mislabel images from a certain manufacturer or hospital’s set of data, then machine learning teams might realize that the image quality from that device isn’t as good as the image quality from other sources or that images taken on a particular day suffered from an odd device setting.
An image from one manufacturer could look very different from the image of another. If they have only 10 percent of images from Siemen devices, they know that they’ll need to gather more Siemen images to ensure that the model can predict well on images captured on that brand of device. The same goes for medical images captured with newer models of devices vs. old ones.
Geographic regions matter, too. Manufacturers tune their devices based on where they’ll be deployed; for instance, contrast settings differ between the US and Europe. Using images from a variety of geographies and manufacturers can help you work against imbuing the machine learning model with bias and to ensure that it generalizes properly.
With the onset of continuous labeling and continuous monitoring, we’ve reached the end of the steps for building a quality assurance workflow for medical image data labeling. The workflow might seem granular in its detail, but a surprising amount of common mistakes occur in medical image annotation when there isn’t a strong workflow in place.
Building a Tool for Medical Annotations Means Understanding Medical Professionals
There are six, equally important steps to structuring a quality assurance workflow for medical image annotation:
- Select and divide the dataset
- Prepare to annotate with multiple blinds
- Establish the labeling protocol
- Practice medical image annotation on a handful of samples
- Release the first batch of images for annotation
- Release the rest of the data for annotation and implement continuous labeling monitoring
However, having the right tools that fit seamlessly into the day-to-day lives of medical professionals is an equally important part of structuring a quality assurance workflow for image annotation.
Encord’s DICOM annotation tool was built in collaboration with clinicians, so it enables medical professionals to navigate and interact with images in the same way that they do in their clinical workflow. We recognize that radiologists and other medical professionals are busy people who have spent years building certain domain expertise and skills.
Our annotation tool mimics and integrates with the clinical experience. Radiologists spend most of their days in dark rooms, using high-resolution grayscale monitors to look at digital images. That’s why our tool supports darkmode, preventing radiologists from encountering a green or white interface during their clinical routine. We also designed a viewer that supports the same method and handling for looking through a volume of image slices, so that they can rely on the muscle memory that they’ve developed through years of using clinical tools.
That’s why we also support hanging protocol. After years of hanging scans up on walls, radiologists are used to seeing them displayed in a certain way. For instance, when reading a mammography, they want to see both breasts at the same time to compare for symmetry and features intrinsic to that particular patient. Rather than ask the radiologist to change for the digital age, we’ve changed the tool to orient images in the way that makes the most sense for their profession.
Our platform, interface, and mouse gestures (including windowing!) were all designed with the clinical experience in mind.
Interested in learning more about Encord’s DICOM Annotation Tool? Reach out for a demo today.
Data infrastructure for multimodal AI
Click around the platform to see the product in action.
Written by

Dr. Andreas Heindl
Explore our products