Announcing our Series C with $110M in total funding. Read more →.
Contents
4 Best Practices for Using DICOM and NIfTI File Format in Computer Vision Models
Encord Blog
How to Annotate DICOM and NIfTI Files
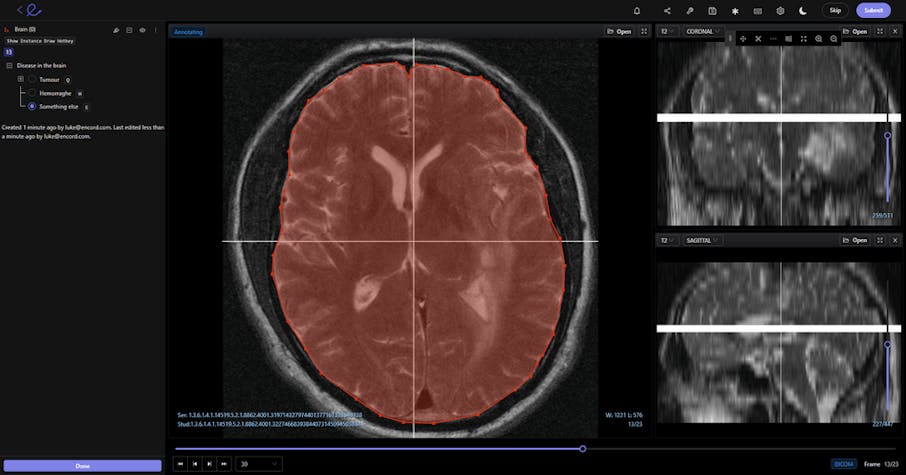
In medical image annotation and computer vision models, the datasets and tools used have highly-specialized requirements. In this article, we will outline the best practice for using DICOM and NIfTI files in computer vision models.
Medical imaging and annotation is a specialized field. Perhaps more so than any others, accuracy is crucial. When we consider the end-users, such as healthcare professionals, and the ultimate outcomes, the impact on patients, we can see why accuracy is crucial.
A correct or incorrect diagnosis impacts treatment, care plans, and outcomes. Accurate computer vision models and machine learning-powered annotation of videos and images can make the difference between life and death for some patients.
Before diving into four best practices to follow when annotating DICOM and NIfTI images, let’s take a moment to consider data security, and clarify how those imaging formats differ from others used, such as PACS and JPEG.
Data Security in Medical Image Labeling and Computer Vision Models
Healthcare providers have strict regulatory oversights worldwide. In every country, government agencies (e.g. the FDA and European CE regulations) and watchdogs carefully monitor compliance of patients’ rights, data protection, and the accuracy of patients’ records.
In the US, for example, HIPAA (the Health Insurance Portability and Accountability Act) is non-negotiable for handling and processing sensitive patient healthcare data, including images, labeling, annotation, and computer vision analysis.
SOC 2 is another benchmark for handling consumer and patient data. It includes an external audit that evaluates data security for the entire end-to-end data process, including computer vision and medical image annotation software. Data security best practices are mission-critical in the healthcare sector.
Medical Imaging Standards Used in Computer Vision Models: DICOM & NIfTI
In most cases, healthcare providers use the DICOM and NIfTI imaging standards. Medical images play a role in that, as 3D and 2D scans — regardless of the imaging standard — are integral to the diagnosis doctors give patients. Accuracy, especially when labeling medical images, is crucial.
Consequently, the software healthcare providers use to label, annotate, and analyze images plays a key role in healthcare decision-making and patient treatment plans. Computer vision models are powerful Artificial Intelligence and Machine Learning-based (AI/ML) software tools for analyzing images.
Healthcare organizations’ commitment to accuracy, excellence, and cost-effective treatment is one of the main drivers behind adopting computer vision models in image and video analysis.
In a previous article, we explained what’s the difference between the DICOM and NIfTI healthcare imaging standards? Have a read to find out more.
What is the difference between the DICOM format and JPEG?
One of the most common imaging formats is JPEG (Joint Photographic Experts Group), and although widely used the world over, it’s not practical or useful in a medical setting. DICOM files contain layers and layers of images, associated metadata, and links to databases and other medical systems.
On the other hand, JPEG files are single-layer 2D images. Medical images in a JPEG format wouldn’t be detailed nor useful enough for medical purposes. Although you can convert DICOM and other files into JPEGs, this is usually convenient when explaining something in simple terms to a patient.
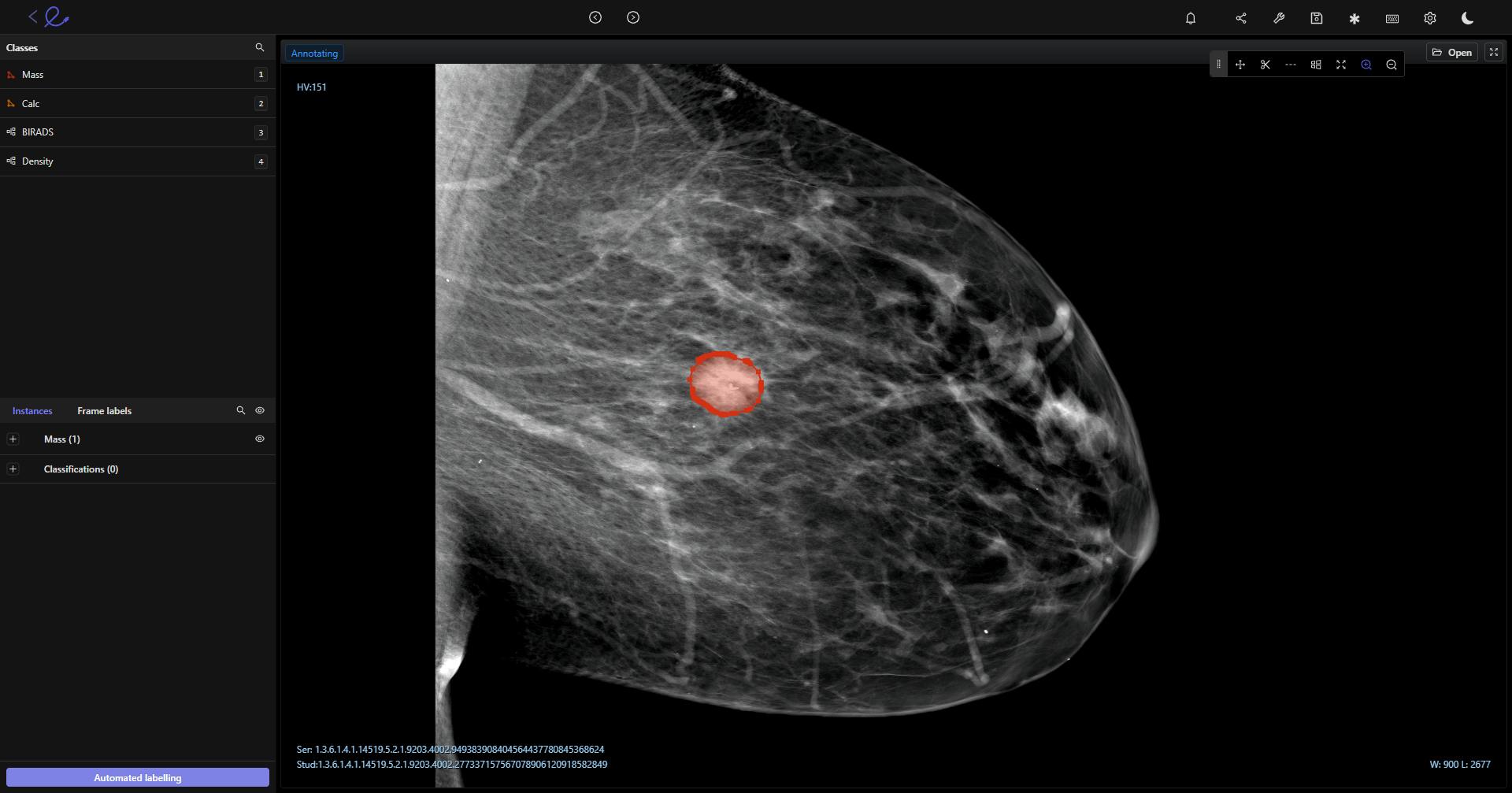
DICOM image being annotated in Encord
What is the difference between DICOM and PACS?
In most healthcare workplaces, doctors and specialists also use the Picture Archiving and Communication System, or PACS, alongside other imaging formats. PACS is used as a medical image storage and archive system, with images being fed into by radiologists and other medical specialists. Images usually come from X-ray machines and MRI scanners.
On the other hand, the DICOM format is an international communication standard for storing, communicating and transmitting medical images with layers of metadata. Medical professionals can use both, with one format supporting the other to ensure every stakeholder involved in patient care has the necessary information.
Now let’s dive into our overview of the four best practices healthcare annotators and medical professionals should apply when using DICOM and NIfTI files in Computer Vision Models.
Compare the different aspects below:
| Aspect | DICOM | JPEG | NIfTI | PACS |
| Purpose | Medical imaging standard for storing and transmitting medical images with metadata | General-purpose image format for 2D images | Specialized format for neuroimaging data | System for storing, archiving, and accessing medical images |
| Data Type | Multi-layered images with detailed metadata (3D & 2D) | Single-layer 2D images | Compact 3D and 4D imaging data | Works with DICOM and other formats for image storage |
| Use Case | CT, MRI, Ultrasound, X-rays; sharing and analyzing images | General photography and basic image sharing | Neuroscience, brain imaging | Image archiving, retrieval, and sharing in healthcare |
| Compression | Lossless or lossy (configurable); optimized for medical use | Lossy (standard); data quality reduced | Typically lossless | Works with compressed/uncompressed images as stored |
| Flexibility | High; supports multiple modalities and workflows | Low; limited to static 2D images | Moderate; designed for specific neuroimaging needs | High; integrates with DICOM and other tools to adapt to various workflows |
| Accessibility | Requires specialized tools for interpretation | Readable by common image viewers | Requires neuroimaging tools for viewing | Accessible via PACS-compatible software |
| Visualization | Suitable for detailed medical analysis; supports layers, slices, and 3D visualization | Basic visualization only | Best for neuroimaging-specific 3D/4D visualizations | Provides access and integration for detailed visualization through DICOM viewers |
| Example Applications | Tumor detection, surgical planning, diagnostic imaging | Patient education, casual sharing | Brain mapping, neuroscience research | Hospital-wide image management, integration with EHR systems |
4 Best Practices for Using DICOM and NIfTI File Format in Computer Vision Models
#1: Display the data correctly to allow for pixel-perfect annotations
When annotators and data scientists talk about displaying the data “correctly”, we mean in a native format. So that when images or videos are uploaded to an AI-based computer vision annotation tool — whether in DICOM format or NIfTI format — nothing is lost.
In the best video annotation and computer vision tools, DICOM or NIfTI formatting is displayed natively. Videos of any length can be displayed and processed.
And crucially, DICOM files come with layers of patient information, such as database connections, image analysis and doctor notes, and even scheduling data for appointments. None of this information must be lost during AI-based computer vision analysis of those images.
- For example, DICOM images are used in CT, MRI, Ultrasound, and RF. Displaying these images in a native format means that annotators and healthcare professionals can accurately measure the size of tumors and other medical problems, and this information can be fed into computer vision models.
- Another example is that displaying gastro and other videos natively means that videos of any length (timescale) can be uploaded, allowing for faster loading, no data loss, and more accurate analysis in computer vision models.
#2: Ensure high levels of medical image annotation quality for computer vision models
Computer vision models need the highest quality of labels and annotation to ensure AI-based algorithms analyze images as accurately as possible.
With the right medical imaging tools and workflows, this is achieved two ways:
- Consensus benchmarks. Having a team of annotators assess and annotate the same images, supported by a computer vision modeling tool, makes achieving a benchmark for expert review easier. Medical imaging is sometimes difficult to assess and not clear-cut, so leveraging multiple experts' consensus helps ensure quality is as high as possible.
- Granular expert review workflows. Medical professionals don’t have time to annotate and label medical images manually. Annotators make the manual inputs; in most cases, those images are processed through computer vision models and tools. After that, annotated images are passed onto an expert, such as a senior radiologist or the company's “annotation gold standard” expert. Any mistakes or inaccuracies are sent back to be re-annotated before the images can be reviewed by healthcare professionals making decisions on patient cases.
#3: Make data audits granular: Mission-critical for healthcare regulatory compliance
Regulatory compliance is mission-critical in healthcare. In the US, healthcare providers have HIPAA, SOC 2, and other data protection laws to consider. In Europe, similar laws and regulations exist, alongside patient and consumer watchdogs and GDPR.
Healthcare providers need the ability to fully audit computer vision model training data, down to the granular level. You should also be able to export data granularly.
Complete control and data auditability ensure no delays or unexpected and unwelcome surprises in the regulatory approval process. This reduces workloads for annotators, admin staff, and departmental heads.
#4: Improve image and video annotation efficiency with automation, to save radiologists valuable time
Annotation work needs to be done as efficiently as possible. Radiologists' and other senior medical professionals’ time is valuable and expensive. Here’s a few ways images can be annotated, labeled and processed efficiently:
A medical imaging tool with an intuitive user-interface. Radiologists are used to working in certain ways, with specific tools and systems (e.g., the Picture Archiving and Communication System, or PACS), and imaging formats, such as DICOM and NIfTI. To ensure radiologists and other medical professionals, any medical imaging tool introduced should have an intuitive interface that’s quick and easy to learn.
Automation features. Manually annotating dozens or hundreds of slides is time-consuming, especially when there’s layers of slides for every DICOM image file. Automation features, such as pre-processing saves hours of manual annotation work, and here’s how this benefits medical professionals and organizations:
- Pre-processing ensures that medical professionals only look at and adjust images that have already been annotated and labeled.
- Automation and machine learning models ensure a higher standard of accuracy, quality, and consistency.
- In microscopy, annotators can manually label cells of interest, and then use automation features to annotate much larger datasets with much greater speed and accuracy.
Encord’s platform is already in use in several healthcare organizations, making positive real-word impacts to patient care. At Stanford Medicine, the Division of Nephrology reduced experiment duration by 80% while processing 3x more images.
Encord has also been deployed in medical image annotation projects by King’s College London and Memorial Sloan Kettering Cancer Center (MSK). Take a look at other case studies here.
Key Takeaways
Before applying computer vision models to DICOM and NIfTI medical images, annotators need to implement several best practice steps to get the best results possible, such as:
- Display the data correctly to allow for pixel-perfect annotations; e.g. using tools that display DICOM and NIfTI in a native format
- Ensure high levels of medical image annotation quality for computer vision models. Manual labeling at the start of the process, and automation tools, generate higher levels of annotation quality and accuracy when images are fed into computer vision models
- Make data audits granular: Mission-critical for healthcare regulatory compliance. With the right tools, you don’t need to worry about this because data granularity and audit-friendly features come as standard
- Improve image and video annotation efficiency with automation, to save radiologists valuable time. Healthcare professionals' time is valuable and expensive. Annotators can save stakeholders and themselves time and money when using automation features, such as pre-processing tools.
Medical image annotation for computer vision models requires accuracy, efficiency, and a high level of quality. With powerful AI-based image annotation tools, medical annotators and professionals can save hours of work and generate more accurately labeled medical images.
Explore the platform
Data infrastructure for multimodal AI
Explore product
Frequently asked questions
JPEG is a single-layer 2D format lacking the metadata and detail necessary for medical analysis. In contrast, DICOM and NIfTI formats provide multi-layered data, enabling in-depth analysis essential for diagnostics and treatment planning.
Data security is critical due to regulations like HIPAA (US) and GDPR (EU). It ensures patient data is handled securely and complies with standards, minimizing risks and maintaining trust.
DICOM is an international communication standard for creating and sharing medical images with associated metadata.
PACS (Picture Archiving and Communication System) is a storage and retrieval system for medical images. Both complement each other to streamline medical imaging workflows.
Computer vision models improve the accuracy and efficiency of image analysis, aiding quicker diagnoses and better treatment planning. They also reduce workloads for annotators and radiologists, saving time and costs.
Yes, users can add custom metadata fields to the files imported into Encord's index. These fields can be tailored to fit specific needs, allowing for categorization into text, numerical, categorical, and date formats. This flexibility enables users to search and filter data effectively based on relevant project requirements.
Encord provides a suite of tools designed to streamline and enhance the annotation process for DICOM healthcare data, including features like the AI segmenter and DICOM editor. These tools help teams manage and curate healthcare data efficiently, ensuring a faster and more accurate annotation workflow.
Encord typically requires band extraction during the preprocessing step for six-band geotiff files. This ensures that the relevant data is correctly formatted and ready for annotation. It's important to provide the full six-band information to facilitate accurate processing.
Encord specializes in supporting clients in the healthcare space with annotation services for DICOM files. Our platform provides the necessary tools to ensure high-quality data annotation, enabling healthcare institutions to leverage AI solutions effectively.
Encord is capable of natively hosting DICOM files and can manage both 2D and 3D data. This includes advanced functionalities like cylindrical brushes for 3D annotation, making it suitable for complex medical imaging and other specialized use cases.
Encord supports various data formats, including DICOM and NIfTI, catering to the needs of different projects. This flexibility allows teams to handle simulation data and other formats effectively, ensuring comprehensive annotation capabilities.
Encord allows users to view Hounsfield units directly on the image while annotating. This feature enhances the annotator's ability to make informed decisions by providing real-time data about the pixel or voxel being analyzed.
Yes, Encord allows users to perform measurements directly within the tool. The platform displays pixel spacing and other relevant metadata attached to DICOM files, enabling accurate measurement of annotated regions.
While Encord does not natively support ECG data, it can process ECG files by converting them into DICOM format for annotation. This flexibility allows users to effectively annotate ECG data within the platform.
Encord's label editor supports various file modalities, including text, audio, images, and video. Users can stack or layout these files in any configuration that suits their evaluation or annotation needs.